Building a Hadoop cluser using Raspberry Pi
Why build this cluster?
Good part:
- Raspberry Pi runs the same Linux and Hadoop as Amazon EC2 does. Good for learning and testing.
- Cheap, less than $200 for the whole system. 40w power consumption, always on 24x7.
- Under same network, no server down, command response has zero lag.
- Versatile, could be multi-machine web server or own cloud.
- Sits on the desk, actually own the server, feels good.
But:
- ARM architecture at low clock speed, slow. Not suited for any big-size dataset.
- However, you can test a little portion of large datasets before shipping to the AWS.
Cluster Physical Setup
Cluster Specs
- 4 x Raspberry Pi 3 with 64 GB SD card
- Gigabit Ethernet Switch for Inter-Nodes talk
- Cables and Power
Assembly
- In order to “stack” the parts together, I ordered a stacking kit which can be stacked together with Raspberry Pi.
- For power supply, I used a 40W 5-Port USB charger with a max output of 2.4A for each port.
Official recommend using a 2.5Amp power supply for each of the Raspberry Pi 3 but I highly doubt they can use that much power. - Since Raspberry Pi 3 has a build-in WIFI adapter, it’s not necessary to use any wired connections in order
to let cluster work. However, for the sake of stability and adaptability, I used a Gigabit Ethernet Switch for inter-nodes communication.
Whenever you need to bring the cluster to somewhere else, you only need to connect the master node to the Internet by wire or WIFI. - The picture of fully assembled cluster: (in the next page due to PDF output)

Cluster Software Setup
Install Linux Image
For Linux distribution, I used RASPBIAN which is official supported operating system. I used RASPBIAN JESSIE LITE which contains no GUI and has only 292 MB.
In order to avoid some stupid mistaks which need several hours to find out, I install the Linux System and Hadoop only into one machine,
configure and test the Hadoop to check if everything is OK and then
dump the whole SD card image to other three of them.
Installation:
- Insert the SD card to my MacBook
diskutil listto list the mounted driverunmount the SD card for writing the image:
diskutil unmountDisk /dev/disk
in my case is
disk3write the image to SD card:
sudo dd bs=1m if=image.img of=/dev/rdisk
and in my case:
sudo dd bs=1m if=/Users/DLI/Downloads/2016-05-27-raspbian-jessie-lite.img of=/dev/rdisk3`
OS setup and Hadoop Compilation
Connect to RPi:
- Insert SD card into one the RPi and connect that RPi to my home router. For clarity, I will name this RPi masterPi.
- Then, log into my router to find the IP address of masterPi,
which is192.168.1.125. Then, manually assign an IP address to masterPi
so I don’t need to check the address of that masterPi after rebooting. log into the
masterPi:SSH pi@192.168.1.125
with a pre-set password:
raspberryexpand the file system and configure the locale setting with
sudo raspi-configdo a
sudo apt-get update && sudo apt-get upgrade
to keep the system fresh
add a new user
HadoopUserinto a sudo grouphadoop:sudo addgroup hadoop sudo adduser --ingroup hadoop HadoopUsershell give me a warning:
adduser: Please enter a username matching the regular expression configured
after some research it seems that Linux doesn’t recommend mixing upper and lower case name,
so I usedsudo adduser --ingroup hadoop hadoopuserinstead (Lession learnt…), then add
hadoopuser to sudo groupsudo adduser hadoopuser sudo
Install JDK
- it is reported that the Oracle JDK has better performance over
Open JDK, so:sudo apt-get install oracle-java8-jdk
note: JDK is installed on/usr/lib/jvm/ check if
java -version
points to Oracle JDK
Download source code
From hadoop-2.7.2
Compile
A quick look at the Building Instruction of Hadoop from Apache, Hadoop has the following requirements:
- Unix System
- JDK 1.6
- Maven 3.0
- Forrest 0.8 (if generating docs)
- Findbugs 1.3.9 (if running findbugs)
- ProtocolBuffer 2.4.1+ (for MapReduce and HDFS)
- CMake 2.6 or newer (if compiling native code)
- Internet connection for first build (to fetch all Maven and Hadoop dependencies)
Addition to JDK, I need at least Maven,ProtocolBuffer and CMake so install them accordingly:
apt-get install maven, a lot of dependencies butapt-getcovered me nicely :)- installing ProtocolBuffer is a little tricky
- we must download the source, then
./autogen.sh, as specified in the offical site by default, ProtocolBuffer install the file to
/usr/localwhich may not be found by the system, so reconfig it to/usr:./configure --prefix=/usrand then
makemake checkto check if builds OK and returns:===================================================== Testsuite summary for Protocol Buffers 3.0.0-beta-3 ===================================================== # TOTAL: 6 # PASS: 6 # SKIP: 0 # XFAIL: 0 # FAIL: 0 # XPASS: 0 # ERROR: 0 =====================================================download the
hadoop-2.7.2source code unzip withtar xzvf hadoop-2.7.2-src.tar.gz
compile the Hadoop natively for RPi
cd hadoop-2.7.2-src/
sudo mvn package -Pdist,native -DskipTests -DtarHowever, I get the error message in the mid of compiling process:
[ERROR] Failed to execute goal org.apache.hadoop:hadoop-maven-plugins:2.7.2:protoc (compile-protoc) on project hadoop-common: org.apache.maven.plugin.MojoExecutionException: ‘protoc –version’ did not return a version -> [Help 1].
After some search online, building Hadoop need version of
protocol buffersexactly be2.5. So I need to delete the one I installed and rebuild Hadoop.- Another 45 minutes of life, I finally get
protocol buffers version 2.5compiled. Again:
sudo mvn package -Pdist,native -DskipTests -Dtar
Finally:
[INFO] ————————————————————————
[INFO] BUILD SUCCESS
[INFO] ————————————————————————
[INFO] Total time: 53:22.700s
[INFO] Finished at: Thu Jun 09 23:08:57 CDT 2016
[INFO] Final Memory: 81M/224M
[INFO] ————————————————————————
Test the build:
bin/hadoop checknative -a
And the output:
Native library checking:
hadoop: true /home/hadoopuser/hadoop-2.7.2-src/hadoop-dist/target/hadoop-2.7.2/lib/native/libhadoop.so.1.0.0
zlib: true /lib/arm-linux-gnueabihf/libz.so.1
snappy: true /usr/lib/libsnappy.so.1
lz4: true revision:99
bzip2: true /lib/arm-linux-gnueabihf/libbz2.so.1
openssl: true /usr/lib/arm-linux-gnueabihf/libcrypto.so
Test with
$ bin/hdfs dfs -put etc/hadoop input
$ bin/hadoop jar share/hadoop/mapreduce/hadoop-mapreduce-examples-2.7.2.jar grep input output 'dfs[a-z.]+'
View the output with
$ bin/hdfs dfs -cat output/*
Output:
6 dfs.audit.logger
4 dfs.class
3 dfs.server.namenode.
2 dfs.period
2 dfs.audit.log.maxfilesize
2 dfs.audit.log.maxbackupindex
1 dfsmetrics.log
1 dfsadmin
1 dfs.servers
1 dfs.replication
1 dfs.file
Everything seems OK, moving to next step:
Clone image to other RPi
Insert the SD card from Pi to my laptop, and then
$ sudo dd if=/dev/sdb of=node1.img
$ sudo dd if=node1.img of=/dev/sdb
Do it 3 times to clone the image to other three slave nodes.
Pseudo-distributed mode test
Setup
In core-site.xml
configuration>
<property>
<name>fs.defaultFS</name>
<value>hdfs://localhost:9000</value>
</property>
</configuration>
And for etc/hadoop/hdfs-site.xml:
<configuration>
<property>
<name>dfs.replication</name>
<value>1</value>
</property>
</configuration>
Then, formate the namenode:
$ bin/hdfs namenode -format
Start the DataNode:
$ sbin/start-dfs.sh
And jps shows:
2608 DataNode
2774 SecondaryNameNode
2520 NameNode
2968 Jps
wordcount test
Download the bioproject.xml from HW1:
wget http://rasinsrv07.cstcis.cti.depaul.edu/CSC555/bioproject.xml
put into HDFS:
$ bin/hdfs dfs -put bioproject.xml bioproject
run wordcount:
root@masterPi:~/hadoop-2.7.2# time bin/hadoop jar
.. share/hadoop/mapreduce/hadoop-mapreduce-examples-2.7.2.jar
.. wordcount bioproject bioprojectOutput2
time report:
real 8m39.367s
user 8m51.510s
sys 0m15.360s
not bad compared to Amazon EC2’s single-node cluster with 1m30s.
Two, Three, Four nodes comparison
two-node cluster
setup
hdfs-site.xml: I lower the block size because of the test sample file size
<configuration>
<property>
<name>dfs.replication</name>
<value>2</value>
</property>
<property>
<name>dfs.blocksize</name>
<value>32m</value>
<description>Block size</description>
</property>
</configuration>
and yarn-site.xml:
then:
bin/hdfs namenode -format
and:
sbin/start-all.sh
Check on the 50070 port:
Node Last contact Admin State Capacity Used Non DFS Used Remaining Blocks
masterPi:50010 (192.168.3.1:50010) 1 In Service 58.61 GB 222.19 MB 9.91 GB 48.48 GB 7
slavePi1:50010 (192.168.3.2:50010) 1 In Service 58.58 GB 222.19 MB 10.44 GB 47.92 GB 7
each of the nodes was allocated 7 blocks.
to start everything and try wordcount on bioproject.xml again.
time bin/hadoop jar share/hadoop/mapreduce/hadoop-mapreduce-examples-2.7.2.jar
wordcount bioproject bioprojectOutput1
the result:
real 8m55.691s
user 8m43.950s
sys 0m15.560s
is not better than the single-node cluster, why? Maybe I changed the blcok size, change it back and re-run the task: now each node has only 2 blocks
result:
real 8m35.001s
user 7m73.950s
sys 0m15.560s
Still the same, why? After some careful observation of the mapreduce job, I see a lot of map -> map jobs which is unrelated to the
mapreduce process. After searching the web, I think the problem is I’m still using the normal hadoop jar ... command which is not
replace by the YARN command that new to the 2.x version of Hadoop.
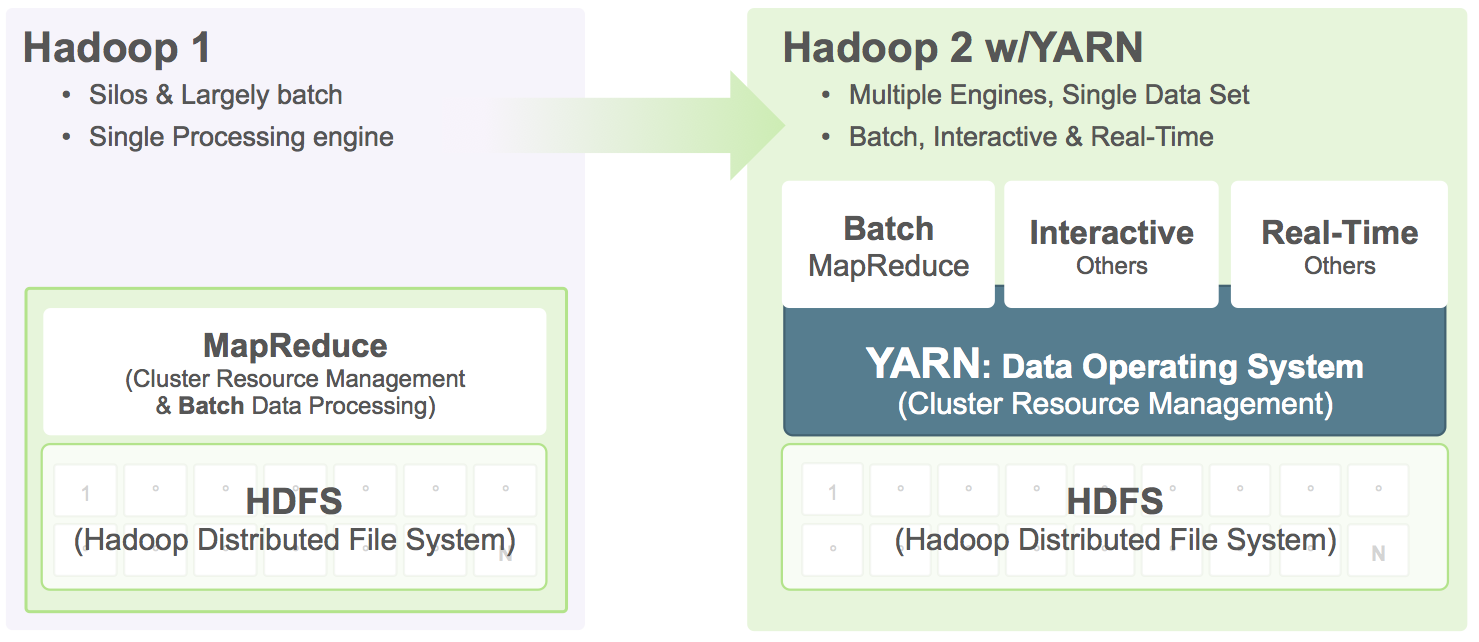
After spending more time doing research on YARN, which is replacing the old ResourceManager by giving authorities to slave node for allocating containers(job runner), monitoring their resource usage.
I find YARN is a great idea since central administration is become harder and harder as the Hadoop ecosystem getting larger. With YARN and MapReduce 2, there are no longer pre-configured static slots for Map and Reduce tasks. The entire cluster is available for dynamic resource allocation of Maps and Reduces as needed by the job. So, I decided to use YARN as task manager instead of Hadoop generic mapreduce. However, after get YARN up and running, I keep getting:
Container killed on request. Exit code is 137
Container exited with a non-zero exit code 137
which is the famous OOM error that the tasks is kill by the system because of not enough memory…
Of course I will get this error message, RPi only has 1g of RAM but YARN will allocate 1024mb of memory minimun for each container and for each node will has multiple containers for the tasks.
So, we must change some parameters:
I added the following in the yarn-set.xml:
<property>
<name>yarn.nodemanager.resource.memory-mb</name>name>
<value>500</value>
</property>
Then, run with command:
bin/yarn jar share/hadoop/mapreduce/hadoop-mapreduce-examples-2.7.2.jar
wordcount /bioproject /bioprojectop1
However, I still get
Diagnostics: Container is running beyond virtual memory limits.
Current usage: 15.1 MB of 256 MB physical memory used; 1.1 GB of 537.6 MB virtual memory used. Killing container.
and containers were shutdown by the system,
Run top shows:
4301 root 20 0 305860 150512 5808 S 70.2 15.1 0:24.66 java
4556 root 20 0 284684 36320 15324 S 62.6 3.6 0:04.16 java
4228 root 20 0 306728 151200 6364 S 61.0 15.2 0:28.99 java
4584 root 20 0 283660 35136 15232 S 60.6 3.5 0:03.36 java
4538 root 20 0 284684 36084 15156 S 59.0 3.6 0:04.15 java
4321 root 20 0 305704 151508 7268 S 56.7 15.2 0:21.70 java
3140 root 20 0 512012 77436 6784 S 17.5 7.8 1:09.14 java
YARN seems give too many works to a single node…
After another servaral hours of configuration, I can’t set parameters properly to let yarn task run successfully. I decided to move on to the next topic and leave this for later to solve.
remove the parameters:
<property>
<name>mapreduce.framework.name</name>
<value>yarn</value>
</property>
in order to bypass the YARN framework and run the Mapreduce tasks directly on HDFS.
Various tasks on fully distributed mode
Add other three nodes to the cluser is just add more names into the slaves file and everything runs smoothly.
wordcount on bioproject
In full-cluster mode, the wordcount job on bioproject give us
real 6m65.001s
user 7m33.050s
sys 0m18.560s
A about 20% decrease in time, not a big increase in performance at all. Run top give us some hint of reason why:
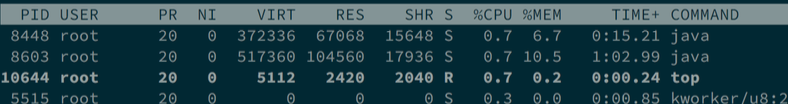
The I/O performance of RPi is so low that CPU is idle waiting for input from the SD card. Moreover, the wordcount is not an CPU but IO intensive task, so the increase in cluster size doesn’t provide too much benefit for RPi.
The bottleneck and improvements
From the result of wordcount on bioproject.xml, we know the bottleneck is from disk IO. In order to optimize it,
we can lower the block size since SD card is not like traditional hardrive which has to spin the disk to read the data.
Also, as indicated by my unsuccessful experiment of
running YARN on top of HDFS, the ResourceManager of YARN could max out the CPU of RPi by spawning many (>10) MapReduce containers to run the tasks in parallel, which I think
could potentially increase the performance. However, containers using too much RAM will get killed by the system. I need to find the sweet spot of balance of parameter setting but I didn’t succeed.
Re-run tasks after optimization
Tweaked the blocksize to 35mb, I run the same project.xml again with result:
real 6m35.833s
user 7m33.343s
sys 0m13.560s
Still, not a big improvement. Search online, I found that we could over-clock the speed of I/O in RPi by modifying the /boot/config.txt
sudo bash -c 'printf "dtoverlay=sdhost,overclock_50=100\n"
>> /boot/config.txt'
Default is 50MHz and I increased to 100. Another run of hadoop:
real 4m52.303s
user 5m23.183s
sys 0m14.560s
Another 20%! I can still push the RPi a little more by overclocking the CPU and RAM speed but I think the increase in performance is limited.
Final thoughts
By moving from disk writing to scheduling of container task which could potentially max out the system resource, the new 2.x Hadoop with YARN architecture is one step to more SPARK like and I think is a elegant solution to the proliferation of Hadoop eco-system.
I tried to configure the YARN parameter correctly but after 2 days of works I still get the out of memory interruption in the middle of task. The reason for that is I still haven’t figured out the multiple relations of different settings. (eg: the minimum size allocated for each container should be larger than adequate but can’t exceed the memory size limit of system)
In conclusion, I learnt a lot from this project. I have a much better understanding of the distributed nature of Hadoop, Linux system and command line environment. Also, presenting my cluster and running demo to the classmates is a fun experience.